Genes¶
The gene entity within the CIViC database includes several useful features for assessing the clinical relevance of variants (Figure 1). The first is a human curated gene-level summary describing the gene’s clinical relevance with associated sources. The second feature is an external link to The Drug Gene Interaction Database, which can be selected to learn more about specific drug-gene interactions and the druggable genome. The third feature is an external link to the ProteinPaint resource allowing a view of variant recurrence data for the gene in the context of alternative transcripts, known protein domains, etc. The fourth feature includes gene-level details (e.g., gene aliases, domains, pathways) pulled in from MyGene.info with a link to additional details.
Understanding Genes
In order to be listed in CIViC a Gene must have at least one clinical Evidence Item that has been curated from the literature and associated with at least one Molecular Profile (variant) of the gene. A new Gene record will be created automatically when the first Evidence Item is assigned to the Gene. The official gene name according to Entrez Gene (assigned by HGNC) is used. Alternative gene names or “aliases” are autopopulated from MyGeneInfo and searchable throughout the database. However, new Evidence Items must be associated with an official gene symbol to prevent ambiguity.
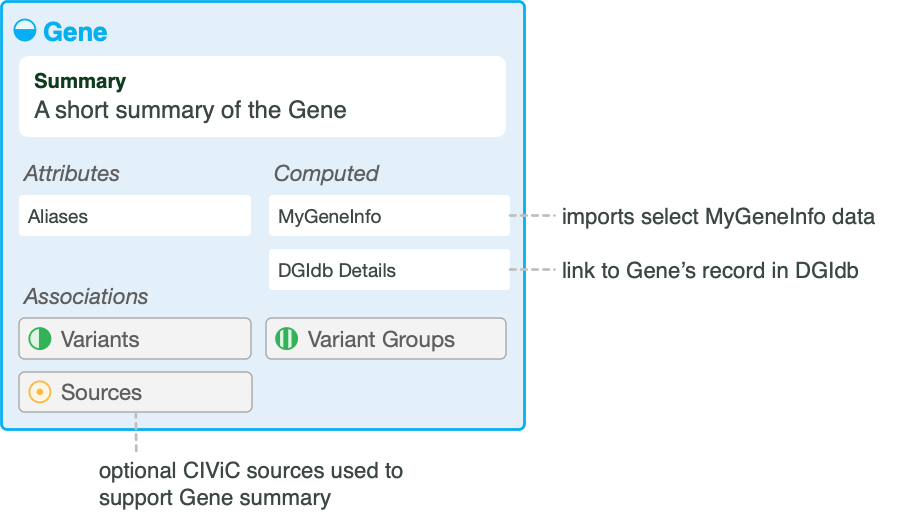
Gene Attributes
Attribute |
Description |
Source |
|---|---|---|
Name |
Entrez symbol of the gene. |
EntrezDB |
Summary |
User-defined summary of the clinical relevance of this Gene. Curation efforts should aim to concisely summarize the relevance of Molecular Profiles (simple or complex variants) in this gene to treatment prediction, prognosis, diagnosis, predisposition, oncogenicity and function. The summary should also provide an overview of the most relevant cancer types. The summary may also include relevant mechanistic information such as pathway interactions, functional alterations caused by variants within this Gene (i.e., activating, loss-of-function, etc.), and normal functions key to its oncogenic properties. |
CIViC |
Sources (PubMed IDs) |
A list of PubMed IDs referring to evidence supporting statements made in the Gene’s description. Source descriptions (e.g. ‘Weisberg et al., 2007, Nat. Rev. Cancer’) are pulled from the PubMed database at the time of submission, and are not editable. |
CIViC (PubMed) |
MyGene.Info |
||
MyGene Info |
Data retrieved from MyGene.Info using the HGNC symbol as the query. Includes synonyms, protein domains, and pathways with additional data displayed by clicking the “Details” button. |
MyGene.Info |
Curating Genes
Most of the information within the Gene entity is automatically imported after the gene is created. These automatically generated fields include: 1) information from MyGene.info and 2) link to DGIdb details, 3) link to ProteinPaint.
Curators can add gene-level summaries and sources associated with the gene-level summaries. These clinical summaries should include relevant cancer subtypes, specific treatments for the gene’s associated variants, pathway interactions, functional alterations caused by the variants in the gene, and normal/abnormal functions of the gene with associated roles in oncogenesis. The sources used for gene-level summaries should correspond to Pubmed IDs.