Variants Overview¶
CIViC variants are usually genomic alterations, including single nucleotide variants (SNVs), insertion/deletion events (indels), copy number alterations (CNVs; such as amplifications or deletions), structural variants (SVs; such as translocations or inversions), and other events that differ from the “normal” genome. In some cases a CIViC variant may represent events of the transcriptome or proteome. For example, ‘expression’ or ‘over-expression’ is a valid variant. New curators should generally avoid proposing new variants that are unlike any already in CIViC. All CIViC Variants are associated with one or more Molecular Profiles with some evidence of clinical relevance (predictive, prognostic, diagnostic, etc).
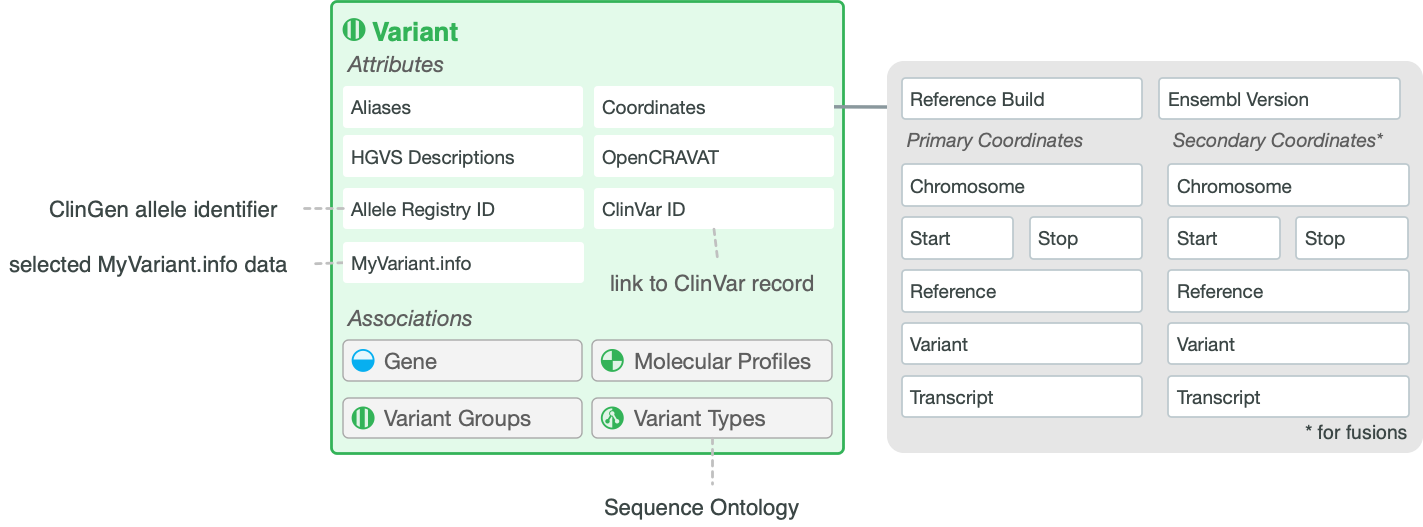
Variant Attributes
Attribute |
Description |
Source |
|---|---|---|
Name |
Description of the type of variant (e.g., ‘V600E’, ‘BCR-ABL fusion’, ‘Loss-of-function’, ‘Exon 12 mutations’). Should be as specific and short as possible (i.e., specific amino acid changes). |
CIViC |
Aliases |
Alternative names for this Variant. May be more verbose (e.g., ‘Val600Glu’) versions, dbSNP IDs or alternative nomenclature used in the literature. |
CIViC |
HGVS Description(s) |
User-defined HGVS strings following HGVS nomenclature that represent this Variant at the DNA, RNA or protein level. |
CIViC |
ClinVar ID(s) |
User-defined ClinVar ID referencing this Variant which will be linked directly to ClinVar. A value of “None Specified” indicates that the variant has not been evaluated for a ClinVar ID. A value of “None Found” indicates that an attempt was made to find a matching ClinVar Entry, but none exists. A value of “N/A” indicates that a ClinVar record is not applicable to the variant (e.g. “Overexpression” variants). |
CIViC (ClinVar) |
Allele Registry ID |
Allele Registry identier (CA id) linked to corresponding ClinGen Allele Registry page. This link is automatically generated using the curated Primary Coordinates (chromosome, start, stop, reference base, variant base). |
ClinGen Allele Registry |
OpenCRAVAT Variant Report |
Link to OpenCRAVAT Variant Report. This link is automatically generated using the curated Primary Coordinates (chromosome, start, stop, reference base, variant base). |
OpenCRAVAT |
Variant Type(s) |
One or more terms from the Sequence Ontology that describes the type of variant. Should be the most descriptive term applicable on a given branch (e.g., ‘Conservative Missense Variant’, ‘Stop Gained’, ‘Transcript Fusion’). |
CIViC (Sequence Ontology) |
Representative Variant Coordinates (Primary Coordinates) |
||
Reference Build |
NCBI or GRC human reference assembly version. |
CIViC |
Ensembl Version |
Ensembl annotation build version. |
CIViC |
Chromosome |
Name of the chromosome in which the variant occurs. |
CIViC |
Start, Stop |
Start and stop positions of the variant (1-based genome coordinate). Start must be less than or equal to stop. |
CIViC |
Reference & Variant Bases |
The nucleotide base of the reference and variant allele (e.g. ‘C’, ‘A’). |
CIViC |
Representative Transcript |
Ensembl transcript ID and version number for a known transcript of the gene that contains the variant (e.g. ‘ENST00000263967.3’). |
CIViC |
Secondary Coordinates |
||
Same as Primary |
For fusion variants, the secondary coordinates are used to specify the 3’ partner of the fusion gene. |
CIViC |
MyVariant.Info |
||
MyVariant Info |
Data retrieved from MyVariant.Info using the curated Primary Coordinates (chromosome, start, stop, reference base, variant base) described above as the query. Includes external IDs and links whenever possible with data displayed in several tabs. |
MyVariant.Info |